生物质能团队夏涛副教授课题组在能源作物和能源微生物的遗传改良与生化转化方面取得多项研究进展
作者:吴雷明何博洋
作物秸秆生物质是极具发展前景的可再生资源,经微生物转化,可以制造大宗生物燃料和高值生物材料。然而,生物质主要来源于植物细胞壁,受细胞壁天然抗降解性和酿酒酵母木糖利用局限性的影响,目前作物秸秆的转化利用存在成本高、效率低等问题。近日,我校生物质与生物能源团队夏涛副教授课题组,整合作物和酵母的遗传改良,以及生物质转化工艺的绿色优化,提出作物秸秆高效绿色资源化利用的新思路。相关研究成果分别以“Down-regulation ofOsMYB103Ldistinctively alters beta-1,4-glucan polymerization and cellulose microfibers assembly for enhanced biomass enzymatic saccharification in rice”和“Double integratingXYL2into engineeredSaccharomyces cerevisiaestrains for consistently enhanced bioethanol production by effective xylose and hexose co-consumption of steam-exploded lignocellulose in bioenergy crops”为题,发表于国际生物能源领域知名期刊Biotechnology for Biofuels和Renewable Energy。
纤维素是生物质的主要成分,其高度结晶化和聚合化是生物质酶解糖化的关键限制因素,通过遗传改良手段,特异性改善纤维素的结构特性是解决这一问题的突破口。夏涛副教授课题组通过鉴定水稻脆秆突变体(Osfc9/myb103),发现下调表达纤维素合成上游转录因子OsMYB103L,显著降低纤维素的结晶指数和聚合度,改变纤维素微纤丝的纳米结构。成熟稻秆经温和预处理后,木质纤维素的酶解糖化效率显著提高10% ~ 28%。此外,RNA-seq和DAP -seq分析发现,OsMYB103L可能通过调控纤维素合酶和微纤丝组装相关基因,介导纤维素的生物合成和有序沉积。该研究为纤维素生物合成的转录调控提供了新思路,有利于能源作物的遗传改良和秸秆的综合利用。

图1.OsMYB103L突变体和转基因植株表型和微纤丝纳米结构观测
传统酿酒酵母的木糖代谢能力低,木糖的有效利用成为木质纤维生物质高效转化的瓶颈问题。已有研究在改良酿酒酵母木糖代谢途径方面开展了大量工作,包括异源表达木糖还原酶XYL1(XR)和木糖醇脱氢酶XYL2(XDH)基因,但由于木糖醇分泌的积累,导致细胞内氧化还原失衡,重组酿酒酵母对木糖发酵缓慢,乙醇产量低。夏涛团队通过筛选合适的融合蛋白连接XDH和XR,同时优化重组菌株的XDH/XR的活性比,达到酿酒酵母高效利用木糖生产乙醇目的。在工业酿酒酵母SF7中,采用基因融合技术表达XYL1和XYL2的重组菌株SF7-Ft3,可以消耗95%木糖。在重组菌株SF7-Ft3过表达XYL2,可持续提高小麦、玉米秸秆和芒草的木糖利用,使生物乙醇产率提高11%-42%。该研究为酵母工程高效利用木糖提供了新策略,并获得2项国家发明专利。
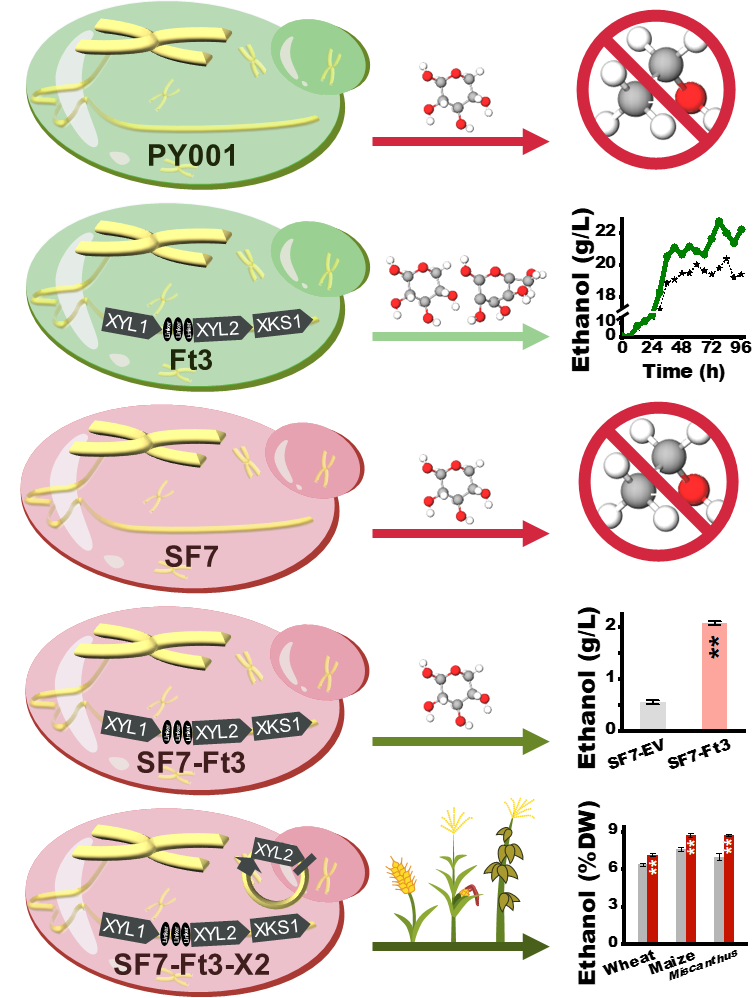
图2.重组酿酒酵母高效利用木糖和葡萄糖生产乙醇的模型图
生科院夏涛副教授为两篇文章的通讯作者,吴雷明博士和张明亮博士为Biotechnology for Biofuels杂志文章的共同第一作者,博士生何博洋为Renewable energy杂志文章第一作者。近年来,夏涛副教授课题组在红外光谱高通量检测细胞壁聚合物技术(Biotechnology for Biofuels,2017)、纤维素结构定向改良与燃料乙醇高效转化机理(Green Chemistry,2018)、木质纤维素绿色解聚与高效酶解机制(Green Chemistry,2019;BioResources,2019)、重金属污染农田作物秸秆无害化处理和纤维素乙醇发酵残渣资源化利用途径(Carbohydrate Polymers,2018;Industrial Crops and Products,2020;Biochemical Engineering Journal,2021)等方面取得了系列研究进展。
Biotechnology for Biofuels摘要:
Background:As a major component of plant cell walls, cellulose provides the most abundant biomass resource convertible for biofuels. Since cellulose crystallinity and polymerization have been characterized as two major features accounting for lignocellulose recalcitrance against biomass enzymatic saccharification, genetic engineering of cellulose biosynthesis is increasingly considered as a promising solution in bioenergy crops. Although several transcription factors have been identified to regulate cellulose biosynthesis and plant cell wall formation, much remains unknown about its potential roles for genetic improvement of lignocellulose recalcitrance.
Results:In this study, we identified a novel rice mutant (Osfc9/myb103) encoded a R2R3-MYB transcription factor, and meanwhile generatedOsMYB103L-RNAi-silenced transgenic lines. We determined significantly reduced cellulose levels with other major wall polymers (hemicellulose, lignin) slightly altered in mature rice straws of themyb103mutant and RNAi line, compared to their wild type (NPB). Notably, the rice mutant and RNAi line were of significantly reduced cellulose features (crystalline index/CrI, degree of polymerization/DP) and distinct cellulose nanofibers assembly. These alterations consequently improved lignocellulose recalcitrance for significantly enhanced biomass enzymatic saccharification by 10%-28% at p < 0.01 levels (n =3) after liquid hot water and chemical (1% H2SO4, 1% NaOH) pretreatments with mature rice straws. In addition, integrated RNA-seq with DAP-seq analyses revealed that theOsMYB103Lmight specifically mediate cellulose biosynthesis and deposition by regulating OsCesAsand other genes associated with microfibril assembly.
Conclusions:This study has demonstrated that down-regulation ofOsMYB103Lcould specifically improve cellulose features and cellulose nanofibers assembly to significantly enhance biomass enzymatic saccharification under green-like and mild chemical pretreatments in rice. It has not only indicated a powerful strategy for genetic modification of plant cell walls in bioenergy crops, but also provided insights into transcriptional regulation of cellulose biosynthesis in plants.
原文链接:
https://doi.org/10.1186/s13068-021-02093-8
Renewable Energy摘要:
Cellulosic ethanol has been regarded as excellent additive into petrol fuels for reduced net carbon release, and yeast fermentation is thus a crucial step for bioethanol production. In this study, three (XYL1/Candida tropicalis, XYL2/Candida tropicalis, XKS1/Saccharomyces cerevisiae) genes were isolated to construct four novel vectors using gene fusion and tandem technology. Four constructs were then transformed into commonSaccharomyces cerevisiaestrain, leading to varied and limited xylose utilization. While two representative constructs were transformed into industrial yeast strain (SF7), the engineered SF7-Ft3 strain could consume 95% of total xylose for ethanol yield at 2.08 g/L, whereas the control strain only utilized 13% xylose with ethanol yield at 0.56 g/L. AdditionalXYL2overexpression into the SF7-Ft3 strain led to consistently enhanced xylose utilization by from diverse enzymatic hydrolats of steam-exploded lignocellulose residues in three major bioenergy crops (wheat, maize,Miscanthus). These consequently increased bioethanol yields (% dry matter) and concentrations (g/L) by 11%-42%. Therefore, this study has demonstrated an applicable yeast-engineering approach for efficient xylose consumption and also provided a powerful strategy for enhancing bioethanol production in bioenergy crops.
原文链接:
https://doi.org/10.1016/j.renene.2021.12.103